
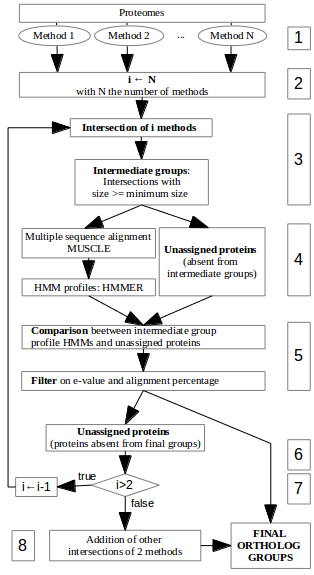
Fig1. MaRiO pipeline overview.
MaRiO is a tool that creates ortholog groups by using existing ortholog groups predictions. MaRiO is the implementation of the meta-approach descripted in the Fig1. It use the result of several methods in order to obtain specific intersections, create relevant profile HMMs and add to this intersections sequences coherent with the profiles.
Publication : Cécile Pereira, Alain Denise and Olivier Lespinet: A meta-approach for improving the prediction and the functional annotation of ortholog groups. BMC Genomics 2014, 16 (Suppl 16)
Then uncompress the tarball (linux: tar -xvzf MaRiO.tar.gz) and please read the ReadMe_MaRiO file.
Fallowing library and programs have to be installed:
The MaRiO program have been positively tested on Linux 64 bts, using hmmer version 3.1b1 and muscle version 3.8.81.
Ortholog groups in fasta and orthoXML format.
In order to evaluate our meta-approach, we checked its consistency according to the ability to predict ortholog groups, and the quality of protein functional annotation within an ortholog group. We used two benchmarks: OrthoBench (first version) and the Orthology Benchmark Service. The values of the parameters used on both benchmark tests for the meta-approach were the same, as stated above: minimum e-value E−10, minimum alignment length of 40%, minimum intersection size equal to four.
The orthologs groups predicts on orthoBENCH data set (involves 1519 proteins from 12 metazoan species, proteome version of Ensembl 72) are available : results on orthobench. File format : protein_id \t group_id
We compared our tool MaRiO with several other common ortholog predictors using the proteome reference dataset and the orthology benchmark service.
The predicted ortholog groups are availables: Mario apply to PRD, file format: specie_id|protein_id \t group_id
Schlicker similarity obtain on orthologs predicted by nine methods plus the meta-approach are available:
For more information, please contact: .