DENSE
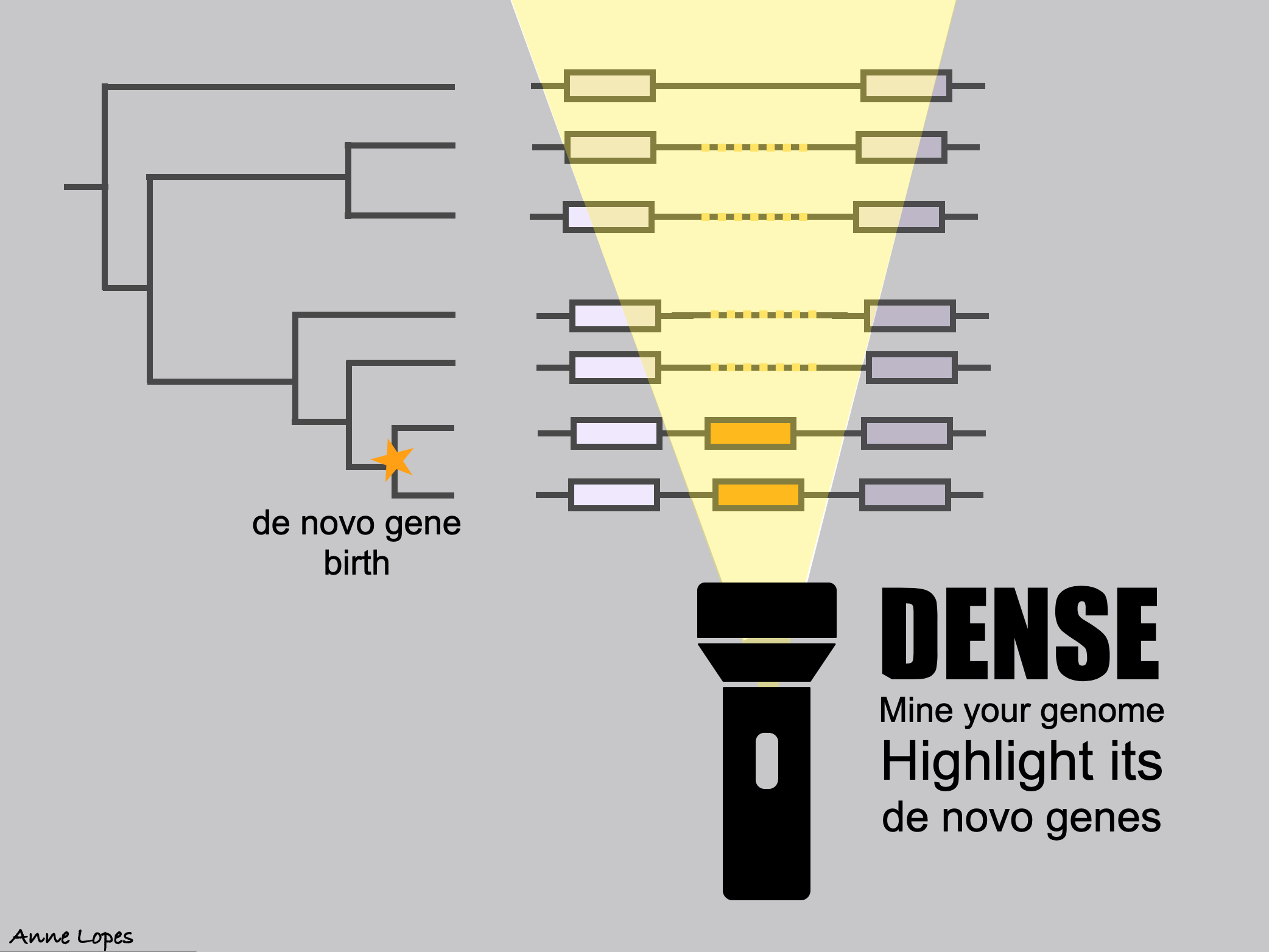
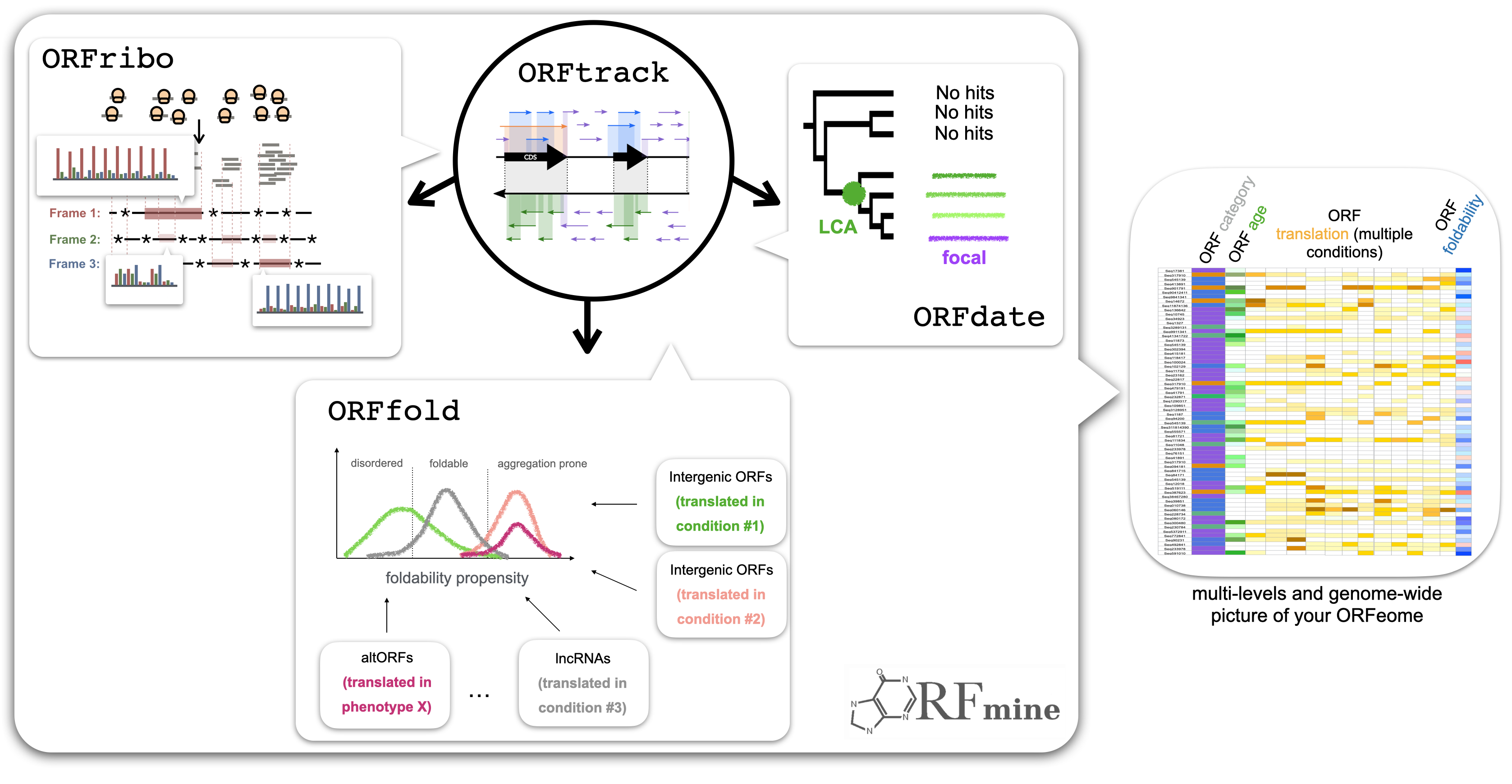
The identification and classification of de novo genes, which originate from noncoding regions of DNA, remain an ongoing challenge in genomic research. While various approaches have been employed for their identification, the lack of a standardized protocol has resulted in varying lists of de novo genes across studies. DENSE (DE Novo emerged gene SEarch) formalizes the common practices used in the field into a comprehensive and automated pipeline. DENSE streamlines the identification of taxonomically restricted genes, homology searches, and synteny analysis. This standardized methodology aims to enhance the accuracy and reliability of de novo gene identification, fostering a deeper understanding of the evolutionary mechanisms that drive gene birth and shape the genetic diversity of organisms. The filtering part of the DENSE workflow, including all criteria combinations, is available through a (web server) once users provide their own list of TRGs. Since calculating the phylostratigraphy can be highly time-consuming, and as many studies involve the same model organisms, we also provide the DENSE calculations for seven model organisms which are compiled into a (public database). These calculations include the phylostratigraphy calculated from the nr database and the predictions obtained from most of the DENSE available combinations of criteria. (Roginski et al, Genome Biology and Evolution 2024).